1. Setup
In a previous post it was shown how to speed up the computation of a kernel density using the Fast Fourier Transform. Conceptually a kernel density is not that far away from kernel regression, accordingly this post is will cover using the FFT to improve the computation of a kernel regression. A popular estimator for the regression function ![]() given observed points
given observed points ![]() is the Nadaraya–Watson estimator with kernel function
is the Nadaraya–Watson estimator with kernel function ![]() and bandwidth
and bandwidth ![]() :
:
(1) 
The FFT requires a grid of with ![]() design points. A meaningful choice in terms of asymptotic is
design points. A meaningful choice in terms of asymptotic is ![]() . Then
. Then ![]() which means that
which means that ![]() and
and ![]() are asymptotically equivalent. In a first step we have to interpolate
are asymptotically equivalent. In a first step we have to interpolate ![]() to fit to an equidistant grid with
to fit to an equidistant grid with ![]() points with
points with
(2) ![]()
Since we use an interpolation using an equidistant grid ![]() (1) becomes
(1) becomes
(3) 
Let the discrete Fourier transform of ![]() be denoted by
be denoted by ![]() . The Fourier transform of (3) for
. The Fourier transform of (3) for ![]() is then using convolution and translation given by
is then using convolution and translation given by
(4) 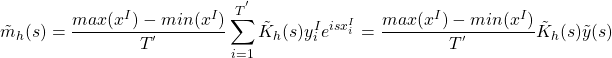
where
2. Asymptotic Considerations
An important question is if the interpolation decline the asymptotic properties of the estimator. Another way to describe the regression setup is for ![]() design points
design points ![]() in addition wlog for simplicity we require
in addition wlog for simplicity we require ![]() there are noisy observations
there are noisy observations ![]() such that
such that
(5) ![]()
for i.i.d. zero mean error terms
![Rendered by QuickLaTeX.com \[\operatorname {E} (({m}(x)-{\hat {m}}(x))^2) \approx \underbrace {h^{4}B^{2}} _{={\text{Bias}}^{2}}+\underbrace {{\frac {1}{Th}}V}_{={\text{Variance}}}}\]](https://www.thebigdatablog.com/wp-content/ql-cache/quicklatex.com-722d9402d4022dd5ae2ae46595a1b567_l3.png)
while using a simple taylor extension, for neigbour points ![]() the error introduced by the interpolation is given by
the error introduced by the interpolation is given by
![]()
Putting all together and using an appropriate bandwidth of order ![]()
![]()
using some straightforward calculations one can show that the interpolating error is dominated by the kernel regression error. A detailed proof and a extension allowing for multivariate ![]() can be found in [2].
can be found in [2].
3. Python Code
To implement the method the fft function that comes with the numpy package was chosen. The numpy fft() function will return the approximation of the DFT from 0 to ![]() . Therefore fftshift() is needed to swap the output vector of the fft() right down the middle. So the output of fftshift(fft()) is then from
. Therefore fftshift() is needed to swap the output vector of the fft() right down the middle. So the output of fftshift(fft()) is then from ![]() to
to ![]() .
.
import numpy as np
import matplotlib.pyplot as plt
import timeit
from scipy.interpolate import interp1d
# Simulation
T = 500
X = np.linspace(-5, 5, T)
Y= (X)**3 + np.random.normal(-0, 10, T)
plt.scatter(X, Y, c="blue")
plt.xlabel('sample')
plt.ylabel('X')
# Presetting
Tout = int( 2 ** np.floor(np.log(T)/np.log(2)) )
print(Tout)
# Interpolate Y
f = interp1d(X, Y)
grid = np.linspace(-5, 5, num=Tout)
Ynew=f(grid)
def epan_kernel(u, b):
u = u / b
return max(0, 1. / b * 3. / 4 * (1 - u ** 2))
# Estimation using FFT
start = timeit.default_timer()
t = grid
h= Tout**(-1/5)
kernel = [epan_kernel(x, h) for x in t]
kernel_ft = np.fft.fft(kernel)
func_tmp = kernel_ft.flatten() * np.fft.fft(Ynew)
m_fft = (max(X)-min(X))*np.fft.fftshift(np.fft.ifft(func_tmp).real)/Tout
plt.plot(t, m_fft)
stop = timeit.default_timer()
print('Time: ', stop - start)
plt.show()
References
[Bibtex]
@book{Fan1996,
added-at = {2009-08-21T10:31:17.000+0200},
address = {London [u.a.]},
author = {Fan, {Jianqing} and Gijbels, {Irène}},
biburl = {http://www.bibsonomy.org/bibtex/23e163e04b09550b54a8067f0cbd97b7e/fbw_hannover},
interhash = {de68bea35adadb13da464f65107efce4},
intrahash = {3e163e04b09550b54a8067f0cbd97b7e},
isbn = {0412983214},
keywords = {Equations Mathematische_Statistik Polynomials Regression_analysis},
number = 66,
pagetotal = {XV, 341},
ppn_gvk = {19282144X},
publisher = {Chapman \& Hall},
series = {Monographs on statistics and applied probability series},
timestamp = {2009-08-21T10:31:17.000+0200},
title = {Local polynomial modelling and its applications},
url = {http://gso.gbv.de/DB=2.1/CMD?ACT=SRCHA&SRT=YOP&IKT=1016&TRM=ppn+19282144X&sourceid=fbw_bibsonomy},
year = 1996
}[Bibtex]
@article{Wand1994,
ISSN = {10618600},
URL = {http://www.jstor.org/stable/1390904},
abstract = {Multivariate extensions of binning techniques for fast computation of kernel estimators are described and examined. Several questions arising from this multivariate extension are addressed. The choice of binning rule is discussed, and it is demonstrated that linear binning leads to substantial accuracy improvements over simple binning. An investigation into the most appropriate means of computing the multivariate discrete convolutions required for binned kernel estimators is also given. The results of an empirical study indicate that, in multivariate settings, the fast Fourier transform offers considerable time savings compared to direct calculation of convolutions.},
author = {M. P. Wand},
journal = {Journal of Computational and Graphical Statistics},
number = {4},
pages = {433--445},
publisher = {[American Statistical Association, Taylor & Francis, Ltd., Institute of Mathematical Statistics, Interface Foundation of America]},
title = {Fast Computation of Multivariate Kernel Estimators},
volume = {3},
year = {1994}
}
Pingback: Efficient Kernel Smoother in the ONNX Format - The Big Data Blog